This git contains all files necessary to reproduce the results of the paper: https://link.springer.com/content/pdf/10.1007%2Fs10994-019-05817-y.pdf
Nuclear discrepancy for single-shot batch active learning
Tom J Viering, Jesse H Krijthe, Marco Loog
in Machine Learning 2019
If you use this code please cite the paper above.
- Clone repository to your local PC:
git clone https://github.com/tomviering/NuclearDiscrepancy.git - Download and put the export_fig package in the folder export_fig under the same folder as the other files.
- Download and put the restools package in the folder restools-master under the same folder as the other files.
- For your convenience, you can find the necessary datasets here: datasets
- I've also included results for MNIST, vehicles and the heart dataset here: results. So you don't need to rerun these experiments and can look at the results.
Note: the results are slightly different than from the ones in the paper, because the random seeds are different. However, the results should not be significantly different.
Runs all experiments. Very time-consuming.
Produces the table with dataset and hyperparameter information:
dataset 01: vehicles, dim: 18, N: 435, pos: 218, sigma: 5.270, lambda -3.0
dataset 02: heart, dim: 13, N: 297, pos: 137, sigma: 5.906, lambda -1.8
dataset 03: sonar, dim: 60, N: 208, pos: 97, sigma: 7.084, lambda -2.6
dataset 04: thyroid, dim: 5, N: 215, pos: 65, sigma: 1.720, lambda -2.6
dataset 05: ringnorm, dim: 20, N: 1000, pos: 510, sigma: 1.778, lambda -3.0
dataset 06: ionosphere, dim: 34, N: 351, pos: 126, sigma: 4.655, lambda -2.2
dataset 07: diabetes, dim: 8, N: 768, pos: 500, sigma: 2.955, lambda -1.4
dataset 08: twonorm, dim: 20, N: 1000, pos: 510, sigma: 5.299, lambda -2.2
dataset 09: banana, dim: 2, N: 1000, pos: 440, sigma: 0.645, lambda -2.2
dataset 10: german, dim: 20, N: 1000, pos: 300, sigma: 4.217, lambda -1.4
dataset 11: splice, dim: 60, N: 1000, pos: 433, sigma: 9.481, lambda -2.6
dataset 12: breast, dim: 9, N: 683, pos: 239, sigma: 4.217, lambda -1.8
dataset 13: mnist3vs5, dim: 784, N: 1000, pos: 484, sigma: 44.215, lambda -6.0
dataset 14: mnist7vs9, dim: 784, N: 1000, pos: 510, sigma: 44.215, lambda -3.6
dataset 15: mnist5vs8, dim: 784, N: 1000, pos: 535, sigma: 44.215, lambda -8.9
Note, lambda here is referring to the regularization parameter, mu = 10^(lambda).
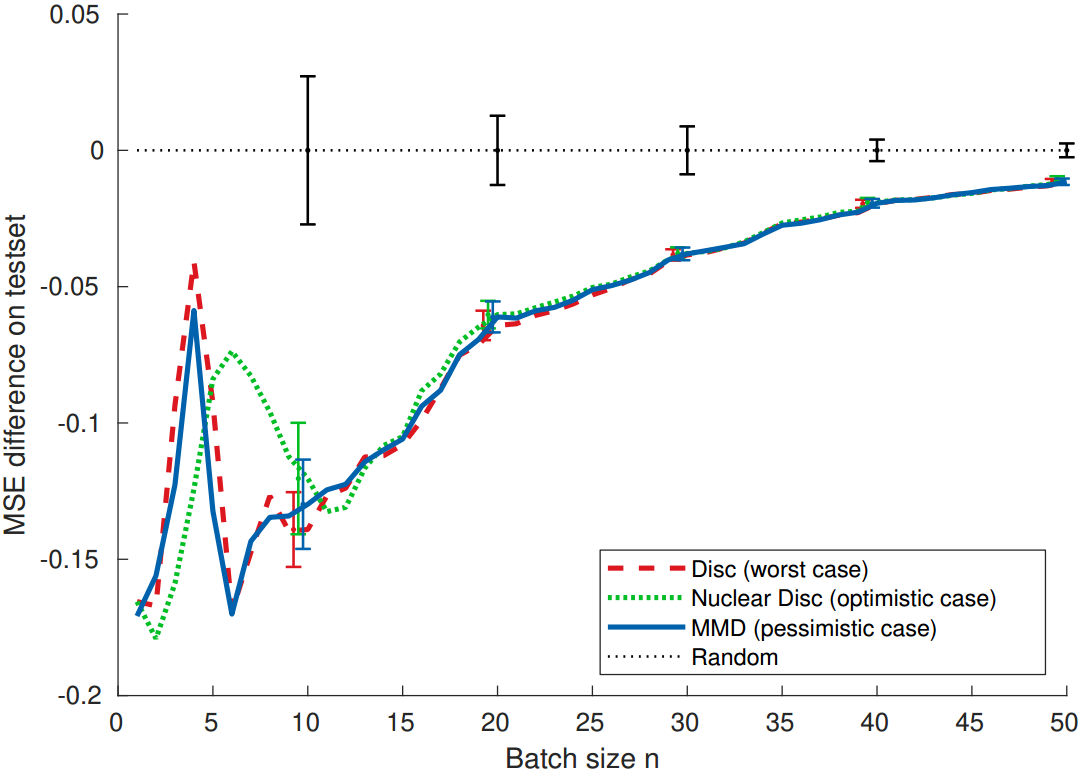
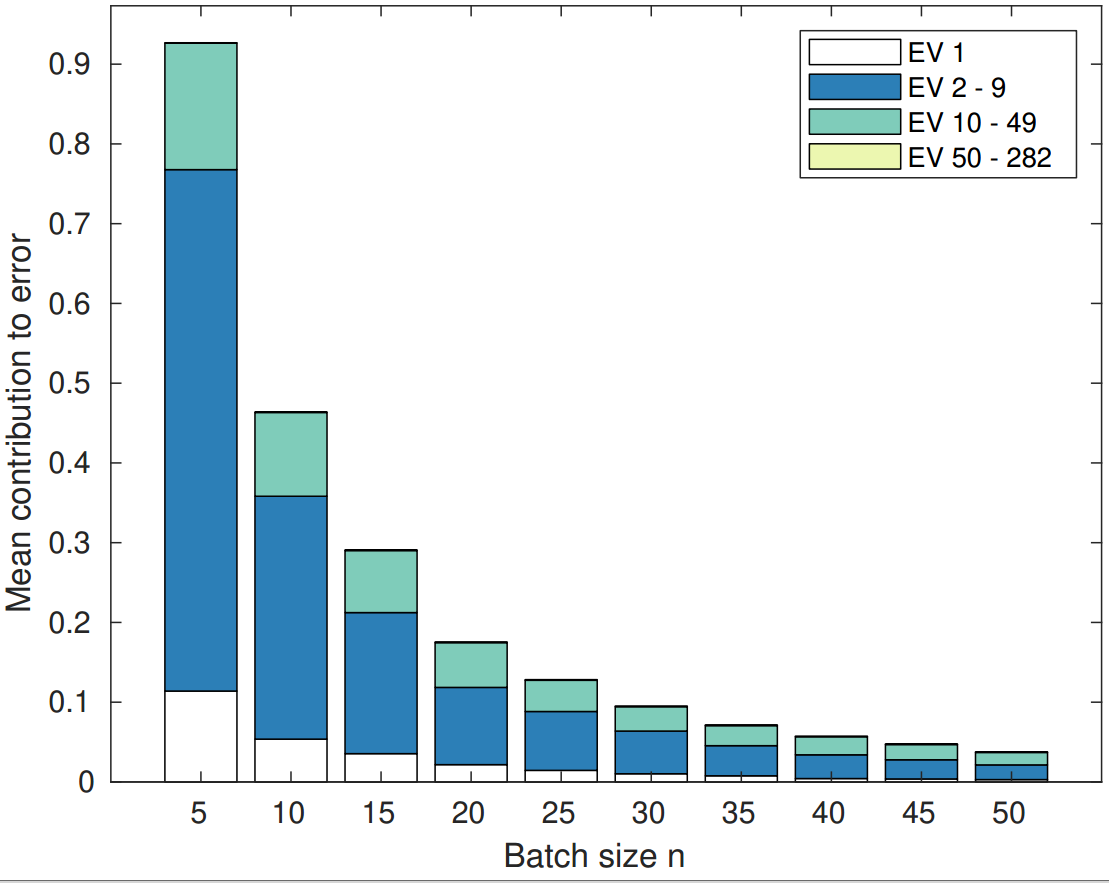
Produces the figures from the paper, such as: learning curves, decomposition of the error.
Produces the table with the Area Under the Learning Curve of the MSE for all datasets. The output is something like:
> **** AULC REA TST *****
**** Average (std over 100) *****
| algorithms
dataset | Random |Discrepancy | MMD |Nuclear Discrepancy
---------+-----------+-----------+-----------+-----------
vehicles | 10.5 (2.4) | 7.3 (1.0) | 7.2 (1.0) | 7.4 (1.1)
heart | 2.9 (0.7) | 2.0 (0.3) | 2.0 (0.4) | 1.8 (0.3)
- 0: random
- 1: crit_mmd: MMD (fast implementation, equation 12)
- 2: crit_mmd2: MMD (slow implementation, equation 6)
- 3: crit_disc: Discrepancy (equation 5)
- 4: crit_ND: Nuclear Discrepancy (first equation of section 5)
If you run into any issues, you can email me: t.j.my_surname_here AT tudelft DOT nl
MIT License
Copyright (c) 2019 T J Viering
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.