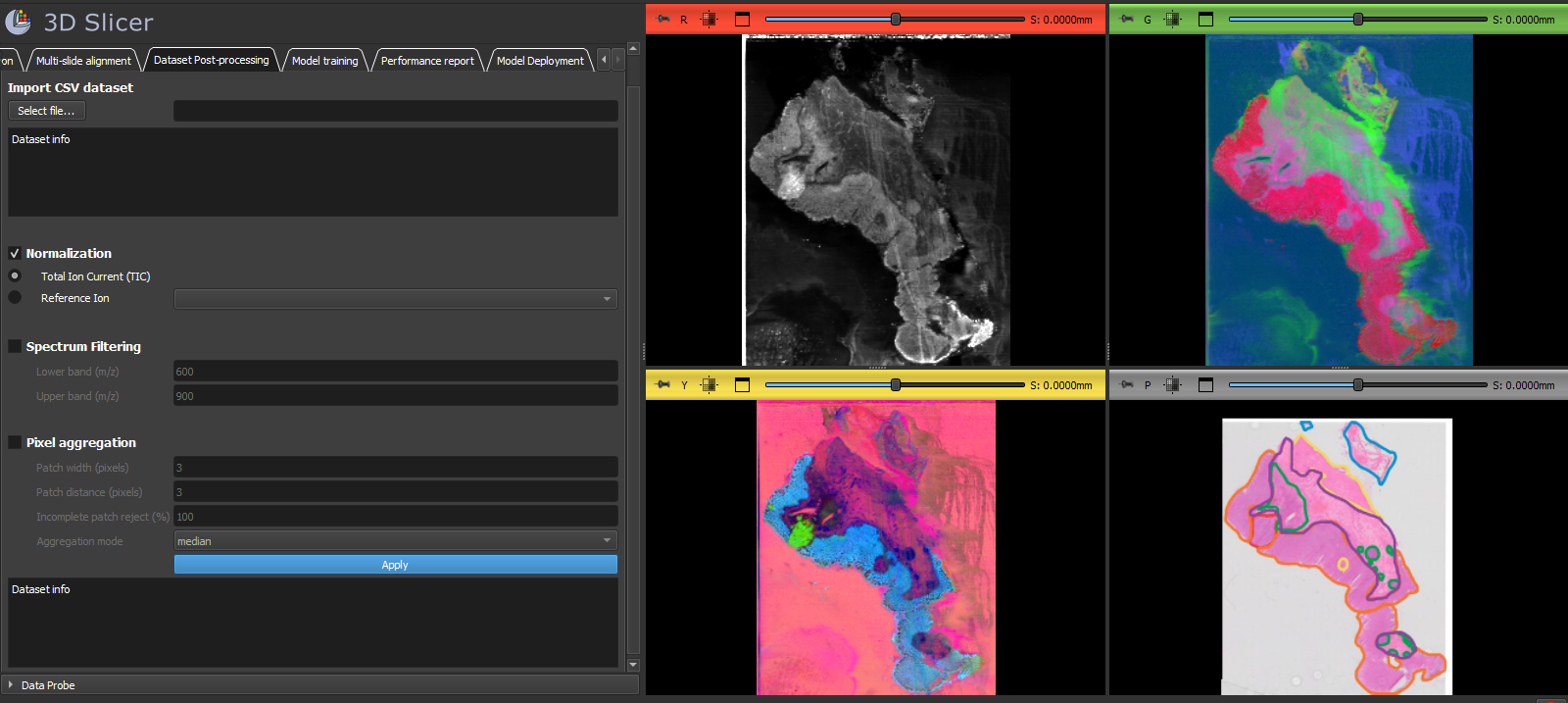
MassVision is a software solution developed in 3D Slicer platform for end-to-end AI-driven analysis of Mass Spectrometry Imaging (MSI) data.
The functionalities include data exploration via various targeted and non-targeted visualization, co-localization to spatial labels (histopathology annotations), dataset curation with spatial- and spectral-guidance, multi-slide dataset merge via feature alignment, denoising via spatial aggregation, AI model training and validation, and whole-slide AI deployment.
- Data format: DESI TXT images, structured CSV MSI
- Visualization: targeted (single-ion heatmap, multi-ion colormap), untargeted (global PCA, regional PCA), pixel spectrum
- Dataset generation: spatial colocalization to pathological annotation, labelled ROI extraction
- Multi-slide merge via feature alignment
- Preprocessing: normalization (TIC, single-ion), subband selection, spatial pixel aggregation
- AI Model training: data stratification, data balancing, model selection
- Whole-slide AI deployment: global deployment, masked deployment
- Visualization with global vs local contrast demo
- Spatial colocalization of mass spectrometry visualization with histopathology annotations demo
- ROI selection with pathology guide for dataset generation demo
Visit here for a complete installation guide from Extentision Manager within 3D Slicer
Visit here to access user guide, tutorials, and other documentations.
MassVision provides users with a test data for exploring and evaluating its functionalities. The test data includes a sample MSI data in a structured CSV format, along with the corresponding histopathology image in TIFF format. The data is available in the release section
Please use the following citations if you use MassVision in your studies and publication
- TBA