-
Notifications
You must be signed in to change notification settings - Fork 41
/
Copy path01-data_visualisation.Rmd
887 lines (678 loc) · 31.2 KB
/
01-data_visualisation.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
500
501
502
503
504
505
506
507
508
509
510
511
512
513
514
515
516
517
518
519
520
521
522
523
524
525
526
527
528
529
530
531
532
533
534
535
536
537
538
539
540
541
542
543
544
545
546
547
548
549
550
551
552
553
554
555
556
557
558
559
560
561
562
563
564
565
566
567
568
569
570
571
572
573
574
575
576
577
578
579
580
581
582
583
584
585
586
587
588
589
590
591
592
593
594
595
596
597
598
599
600
601
602
603
604
605
606
607
608
609
610
611
612
613
614
615
616
617
618
619
620
621
622
623
624
625
626
627
628
629
630
631
632
633
634
635
636
637
638
639
640
641
642
643
644
645
646
647
648
649
650
651
652
653
654
655
656
657
658
659
660
661
662
663
664
665
666
667
668
669
670
671
672
673
674
675
676
677
678
679
680
681
682
683
684
685
686
687
688
689
690
691
692
693
694
695
696
697
698
699
700
701
702
703
704
705
706
707
708
709
710
711
712
713
714
715
716
717
718
719
720
721
722
723
724
725
726
727
728
729
730
731
732
733
734
735
736
737
738
739
740
741
742
743
744
745
746
747
748
749
750
751
752
753
754
755
756
757
758
759
760
761
762
763
764
765
766
767
768
769
770
771
772
773
774
775
776
777
778
779
780
781
782
783
784
785
786
787
788
789
790
791
792
793
794
795
796
797
798
799
800
801
802
803
804
805
806
807
808
809
810
811
812
813
814
815
816
817
818
819
820
821
822
823
824
825
826
827
828
829
830
831
832
833
834
835
836
837
838
839
840
841
842
843
844
845
846
847
848
849
850
851
852
853
854
855
856
857
858
859
860
861
862
863
864
865
866
867
868
869
870
871
872
873
874
875
876
877
878
879
880
881
882
883
884
885
886
887
# (PART\*) Whole game {-}
# Data visualisation
**Learning objectives:**
:::: {style="display: flex;"}
::: {}
- Produce a simple plot with `ggplot2`
- Use different **`aes`thetic mappings** and **`geom_*()` functions** to produce more complex plots
- Visualize **distributions and relationships** in data
- Produce **small multiples** with `facet()`
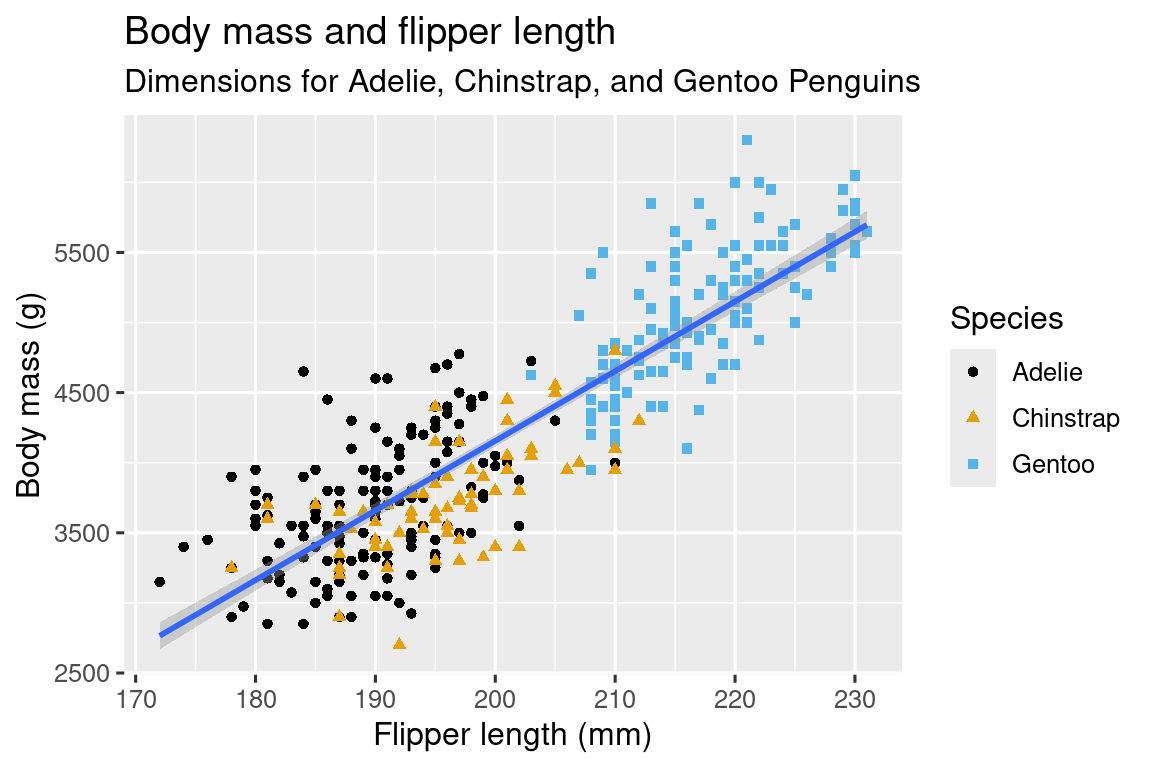
- [Make the plot to the right](https://r4ds.hadley.nz/data-visualize_files/figure-html/unnamed-chunk-7-1.png)
:::
::: {}
:::
::::
## Loading Packages in R {-}
- `install.packages("Package_name")` to **install** a package.
- Before 1st use, but only once
- Packages you need may already be installed
- `library(Package_name)` to **load** a package.
- In general, do this at start of session
- Alternatively: `ggplot2::ggplot()`.
- "ggplot2" = package, "ggplot()" = function
```{r 02-library}
library(tidyverse)
```
We'll also need...
```{r 02-otherpkgs, message=FALSE, warning=FALSE}
library(palmerpenguins)
library(ggthemes)
```
## Peeking at the `palmerpenguins` {-}
```{r 02-dataset}
palmerpenguins::penguins
```
## Creating a ggplot: ggplot() {-}
- Call `ggplot()` function.
- Arguments:
- **data** to use
- how to **map** them to **aesthetics**
```{r 02-plot01, message=FALSE, warning=FALSE}
ggplot(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g)
)
```
## Creating a ggplot: geom_\*() {-}
```{r 02-plot02}
ggplot(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g)
) +
geom_point()
```
(warning = 2 penguins in dataset w/ missing body mass / flipper length)
## Adding Aesthetics and Layers {-}
- **Aesthetics** = visual properties of the plot.
- More aesthetics may tell the data's story better.
- Types of **aesthetic mapping**:
- coordinates: x and y
- linewidth
- shape
- color
- fill
- alpha (transparency)
- stroke
- linetype
- group
- show.legend
- others, sometimes specific to geom
```{r 02-plot03, message=FALSE, warning=FALSE}
ggplot(
data = penguins,
mapping = aes(
x = flipper_length_mm,
y = body_mass_g,
color = species
)
) +
geom_point()
```
## Adding Layers {-}
- Additional `geom_*()` layers may help as well
```{r 02-plot04, message=FALSE, warning=FALSE}
ggplot(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g, color = species)
) +
geom_point() +
geom_smooth(method = "lm")
```
- `geom_*()` functions available:
- point (scatterplot)
- line
- smooth (curve)
- histogram / bar or (stat_count) / col
- boxplot
- map
- text / label
- ...
## Global vs. Local Aesthetics {-}
:::: {style="display: flex;"}
::: {style="margin-right: 0.5em; width: 50%;"}
**Global aesthetics:**
- in `ggplot()`
- apply to all layers
```{r 02-plot05, message=FALSE, warning=FALSE}
ggplot(
data = penguins,
mapping = aes(
x = flipper_length_mm,
y = body_mass_g,
color = species
)
) +
geom_point() +
geom_smooth(method = "lm")
```
:::
::: {style="margin-left: 0.5em; width: 50%;"}
**Local aesthetics:**
- in `geom_*()`
- apply only to that layer.
```{r 02-plot06, message=FALSE, warning=FALSE}
ggplot(
data = penguins,
mapping = aes(
x = flipper_length_mm,
y = body_mass_g
)
) +
geom_point(
aes(color = species)) +
geom_smooth(method = "lm")
```
:::
::::
## Improving Accessibility {-}
- Mapping shape and color may help colorblind readers
- Colorblind-friendly palette would help, too
```{r 02-plot07, message=FALSE, warning=FALSE}
ggplot(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g)
) +
geom_point(aes(color = species, shape = species)) +
geom_smooth(method = "lm") +
scale_color_colorblind()
```
## Why Stop There? {-}
Use `labs()` to add understandable titles and axes.
```{r 02-plot08, message=FALSE, warning=FALSE}
ggplot(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g)
) +
geom_point(aes(color = species, shape = species)) +
geom_smooth(method = "lm") +
labs(
title = "Body mass and flipper length",
subtitle = "Dimensions for Adelie, Chinstrap, and Gentoo Penguins",
x = "Flipper length (mm)", y = "Body mass (g)",
color = "Species", shape = "Species"
) +
scale_color_colorblind()
```
## Exercises {-}
**1. How many rows are in `penguins`? How many columns?**
```{r 02-ex01-01}
penguins
```
See first line. 344 rows x 8 columns.
**2. What does the `bill_depth_mm` variable in the `penguins` data frame describe? Read the help for `?penguins` to find out.**
Call `?penguins` for definition from package documentation
**3. Make a scatterplot of `bill_depth_mm` vs. `bill_length_mm`. That is, make a scatterplot with `bill_depth_mm` on the y-axis and `bill_length_mm` on the x-axis. Describe the relationship between these two variables.**
```{r 02-ex03-01, message=FALSE, warning=FALSE}
ggplot(penguins) +
geom_point(aes(x = bill_depth_mm, y = bill_length_mm))
```
Positive, linear relationship? We'll see more about this in later exercises.
**4. What happens if you make a scatterplot of species vs. bill_depth_mm? What might be a better choice of geom?**
```{r 02-ex04-01}
ggplot(penguins) +
geom_point(aes(x = species, y = bill_depth_mm))
```
Dotplot for every species. Boxplot would tell us more.
```{r 02-ex04-02}
ggplot(penguins) +
geom_boxplot(aes(x = species, y = bill_depth_mm))
```
This better compares bill depths across species.
**5. Why does the following give an error and how would you fix it?**
```{r 02-ex05-01, eval=FALSE}
ggplot(data = penguins) +
geom_point()
```
The error reads `! geom_point() requires the following missing aesthetics: x and y`. Call needs x variable and y variable.
**6. What does the `na.rm` argument do in `geom_point()`? What is the default value of the argument? Create a scatterplot where you successfully use this argument set to `TRUE`.**
From function documentation, "If `FALSE`, the default, missing values are removed with a warning. If `TRUE`, missing values are silently removed."
```{r 02-ex06-01}
ggplot(penguins) +
geom_point(aes(x = species, y = bill_depth_mm), na.rm = TRUE)
```
**7. Add the following caption to the plot you made in the previous exercise: "Data come from the `palmerpenguins` package." Hint: Take a look at the documentation for `labs()`.**
`caption` argument adds a caption.
```{r 02-ex07-01, message=FALSE, warning=FALSE}
ggplot(penguins) +
geom_boxplot(aes(x = species, y = bill_depth_mm)) +
labs(
title = "Distribution of Penguin Bill Depths by Species",
subtitle = "For penguins at Palmer Station Antarctica",
x = "Species",
y = "Bill depth (in millimeters)",
caption = "Data come from the `palmerpenguins` package."
)
```
**8.Recreate the following visualization. What aesthetic should `bill_depth_mm` be mapped to? And should it be mapped at the global level or at the geom level?**
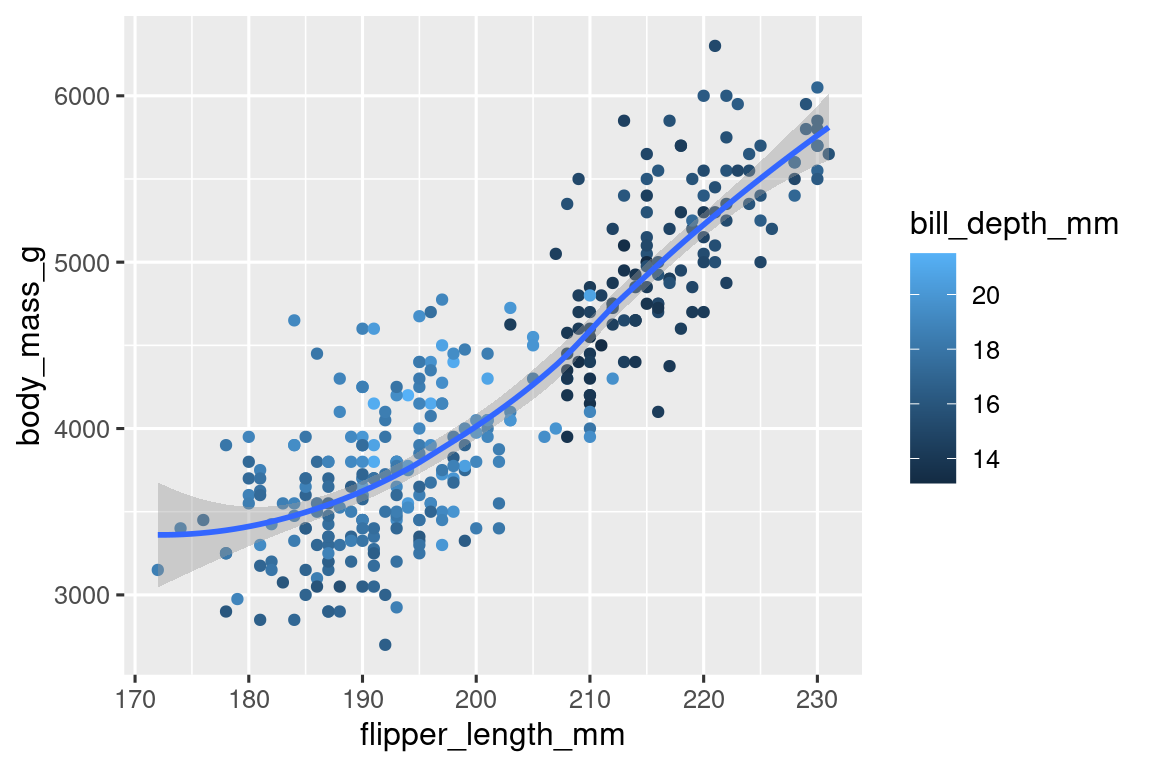
```{r 02-ex08-01, message=FALSE, warning=FALSE}
ggplot(data = penguins, aes(x = flipper_length_mm, y = body_mass_g)) +
geom_point(aes(color = bill_depth_mm)) +
geom_smooth(method = "loess")
```
**9. Run this code in your head and predict what the output will look like. Then, run the code in R and check your predictions.**
```{r 02-ex09-02, message=FALSE, warning=FALSE}
ggplot(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g, color = island)
) +
geom_point() +
geom_smooth(se = FALSE)
# THINK BEFORE SCROLLING DOWN
#
#
#
#
#
#
#
```
**10. Will these two graphs look different? Why/why not?**
```{r 02-ex10-01, eval=FALSE, message=FALSE, warning=FALSE}
ggplot(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g)
) +
geom_point() +
geom_smooth()
ggplot() +
geom_point(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g)
) +
geom_smooth(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g)
)
```
Only difference is mapping globally vs. locally, and mapped same in both geoms, so they should look the same.
```{r 02-ex10-02, echo=FALSE, message=FALSE, warning=FALSE}
ggplot(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g)
) +
geom_point() +
geom_smooth()
ggplot() +
geom_point(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g)
) +
geom_smooth(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g)
)
```
## `ggplot2` calls {-}
- For brevity, "data =" and "mapping =" often dropped
- But things still work...
```{r 02-twin02, message=FALSE, warning=FALSE}
ggplot(penguins, aes(x = flipper_length_mm, y = body_mass_g)) +
geom_point()
```
## Visualizing Distributions: Barplots {-}
Quickly check out variable distributions.
Barplots help with categorical variables.
```{r 02-categorical, message=FALSE, warning=FALSE}
ggplot(penguins, aes(x = species)) +
geom_bar()
```
## Visualizing Distributions: Histograms {-}
Histograms aid with numerical variables
```{r 02-numerical, message=FALSE, warning=FALSE}
ggplot(penguins, aes(x = body_mass_g)) +
geom_histogram(binwidth = 200)
```
## Visualizing Distributions: Density plots {-}
Density plots also help
```{r 02-numerical2, message=FALSE, warning=FALSE}
ggplot(penguins, aes(x = body_mass_g)) +
geom_density()
```
## (more) Exercises {-}
**1. Make a bar plot of species of penguins, where you assign species to the y aesthetic. How is this plot different?**
```{r 02-ex11, message=FALSE, warning=FALSE}
ggplot(penguins, aes(y = species)) +
geom_bar()
```
Bars horizontal instead of vertical.
**2. How are the following two plots different? Which `aesthetic`, `color` or `fill`, is more useful for changing the color of bars?**
```{r 02-ex12, message=FALSE, warning=FALSE}
ggplot(penguins, aes(x = species)) +
geom_bar(color = "red")
ggplot(penguins, aes(x = species)) +
geom_bar(fill = "red")
```
For `geom_bar()`, "color" affects borders. "Fill" affects inside.
**3. What does the `bins` argument in geom_histogram() do?**
The `bins` argument is helpful when you don't have a particular bin width in mind, but you do want to narrow things down to a particular number of bins.
**4. Make a histogram of the `carat` variable in the `diamonds` dataset that is available when you load the tidyverse package. Experiment with different binwidths. What binwidth reveals the most interesting patterns?**
Let's try 1, 0.1, and 0.01.
```{r 02-ex14-01, message=FALSE, warning=FALSE}
ggplot(diamonds) +
geom_histogram(aes(x = carat), binwidth = 1)
```
```{r 02-ex14-02, message=FALSE, warning=FALSE}
ggplot(diamonds) +
geom_histogram(aes(x = carat), binwidth = 0.1)
```
```{r 02-ex14-03, message=FALSE, warning=FALSE}
ggplot(diamonds) +
geom_histogram(aes(x = carat), binwidth = 0.01)
```
Here, a `binwidth` of 0.1 seems to be a good compromise between showing granularity in the data and not overwhelming ourselves with too many bars.
## Visualizing relationships {-}
`ggplot2` is also incredible at displaying relationships between variables.
## Numerical Variable and Categorical Variable {-}
Boxplots help here (or color-coded density plots).
```{r 02-relationship-boxplot, message=FALSE, warning=FALSE}
ggplot(penguins, aes(x = species, y = body_mass_g)) +
geom_boxplot()
```
## Two Categorical Variables {-}
Try a bar plot.
```{r 02-relationship-bar, message=FALSE, warning=FALSE}
ggplot(penguins, aes(x = island, fill = species)) +
geom_bar()
```
To get proportions, set `position` argument to "fill"
```{r 02-relationship-barprop, message=FALSE, warning=FALSE}
ggplot(penguins, aes(x = island, fill = species)) +
geom_bar(position = "fill")
```
## Two Numerical Variables {-}
Scatterplots shine here.
```{r 02-relationship-scatter, message=FALSE, warning=FALSE}
ggplot(penguins, aes(x = flipper_length_mm, y = body_mass_g)) +
geom_point()
```
## Three or More Variables {-}
Map each to different variable
```{r 02-relationship-scatter-multi, message=FALSE, warning=FALSE}
ggplot(penguins, aes(x = flipper_length_mm, y = body_mass_g)) +
geom_point(aes(color = species, shape = island))
```
Or use `facet_wrap()` to create "small multiples"
```{r 02-relationship-facet, message=FALSE, warning=FALSE}
ggplot(penguins, aes(x = flipper_length_mm, y = body_mass_g)) +
geom_point(aes(color = species, shape = species)) +
facet_wrap(~island)
```
## (Even More) Exercises {-}
**1. The `mpg` data frame that is bundled with the `ggplot2` package contains 234 observations collected by the US Environmental Protection Agency on 38 car models. Which variables in `mpg` are categorical? Which variables are numerical? (Hint: Type `?mpg` to read the documentation for the dataset.) How can you see this information when you run `mpg?`**
Running `glimpse(mpg)` gets us most of the way there. We can assume all of the character-based variables ("\<chr\>") are categorical. Since we know we're dealing with cars, we can also recognize that year and cyl (number of cylinders), while technically numerical, are effectively categorical as there is a limited number of possible results. That leaves displ (engine displacement), cty (mpg in city), and hwy (mpg on a highway) as purely numerical.
**2. Make a scatterplot of `hwy` vs. `displ` using the `mpg` data frame. Next, map a third, numerical variable to color, then size, then both color and size, then shape. How do these aesthetics behave differently for categorical vs. numerical variables?**
```{r 02-ex16-01, message=FALSE, warning=FALSE}
ggplot(mpg) +
geom_point(aes(x = hwy, y = displ))
```
```{r 02-ex16-02, message=FALSE, warning=FALSE}
ggplot(mpg) +
geom_point(aes(x = hwy, y = displ, color = cty))
```
```{r 02-ex16-03, message=FALSE, warning=FALSE}
ggplot(mpg) +
geom_point(aes(x = hwy, y = displ, size = cty))
```
```{r 02-ex16-04, message=FALSE, warning=FALSE}
ggplot(mpg) +
geom_point(aes(x = hwy, y = displ, color = cty, size = year))
```
```{r 02-ex16-05}
ggplot(mpg) +
geom_point(aes(x = hwy, y = displ, color = cty, size = year, shape = manufacturer))
```
First off, shape doesn't accept numerical variables. When a categorical variable is mapped to size, a warning suggests this is ill-advised. With color, numerical variables are given a gradient palette while categoricals get varying colors (We've seen this behavior before [think penguin species], so it's not plotted here).
**3. In the scatterplot of `hwy` vs. `displ`, what happens if you map a third variable to `linewidth`?**
```{r 02-ex17-01}
ggplot(mpg) +
geom_point(aes(x = hwy, y = displ, color = cty, size = year, linewidth = manufacturer))
```
It's ignored because there are no lines in a scatterplot. (Also, if you use a categorical variable, it advises you against it).
**4. What happens if you map the same variable to multiple aesthetics?**
```{r 02-ex18-01, message=FALSE, warning=FALSE}
ggplot(mpg) +
geom_point(aes(x = hwy, y = displ, color = cty, size = cty))
```
Both work, but it can be kind of redundant. It can be useful for colorblind users, though (as we saw before with color + shape).
**5. Make a scatterplot of `bill_depth_mm` vs. `bill_length_mm` and color the points by `species`. What does adding coloring by species reveal about the relationship between these two variables? What about faceting by species?**
```{r 02-ex19-01, message=FALSE, warning=FALSE}
ggplot(penguins) +
geom_point(aes(x = bill_depth_mm, y = bill_length_mm, color = species))
```
```{r 02-ex19-02, message=FALSE, warning=FALSE}
ggplot(penguins) +
geom_point(aes(x = bill_depth_mm, y = bill_length_mm, color = species))+
facet_wrap(~species)
```
Color-coding helps spot the relationship of bill sizes by species (compare to Exercise 3 in the first block of exercises). Faceting helps the eye notice those relationships a little better by taking the "noise" of the other species out of the picture.
**6. Why does the following yield two separate legends? How would you fix it to combine the two legends?**
```{r 02-ex20-01}
ggplot(
data = penguins,
mapping = aes(
x = bill_length_mm, y = bill_depth_mm,
color = species, shape = species
)
) +
geom_point() +
labs(color = "Species")
```
Adding an argument to `labs` about our shape as well as our color should solve that.
```{r 02-ex20-02, message=FALSE, warning=FALSE}
ggplot(
data = penguins,
mapping = aes(
x = bill_length_mm, y = bill_depth_mm
)
) +
geom_point(aes(color = species, shape = species)) +
labs(color = "Species", shape = "Species")
```
**7. Create the two following stacked bar plots. Which question can you answer with the first one? Which question can you answer with the second one?**
```{r 02-ex21-01, message=FALSE, warning=FALSE}
ggplot(penguins, aes(x = island, fill = species)) +
geom_bar(position = "fill")
ggplot(penguins, aes(x = species, fill = island)) +
geom_bar(position = "fill")
```
The first tells us which species make up certain proportions of an island's overall population. The second tells us how a species' overall population is split among the islands.
## Saving Your Plots {-}
`ggsave()` lets you get your plots out of R for sharing.
```{r 02-ggsave, eval=FALSE}
ggplot(penguins, aes(x = flipper_length_mm, y = body_mass_g)) +
geom_point()
ggsave(filename = "penguin-plot.png", width = 800, units = "px")
#> Saving 800 x 1983 px image
#> Warning message:
#> Removed 2 rows containing missing values (`geom_point()`).
```
Setting `width` and/or `height` helps with reproducibility.
## (Yet More) Exercises {-}
**1. Run the following lines of code. Which of the two plots is saved as `mpg-plot.png`? Why?**
```{r 02-ex22-01, warning=FALSE, message=FALSE, eval=FALSE}
ggplot(mpg, aes(x = class)) +
geom_bar()
ggplot(mpg, aes(x = cty, y = hwy)) +
geom_point()
ggsave("mpg-plot.png")
```
It will run the second. In looking at `?ggplot2::ggsave`, we see that it defaults to the most recent plot run.
**2. What do you need to change in the code above to save the plot as a PDF instead of a PNG? How could you find out what types of image files would work in `ggsave()`?**
Changing filename in `ggsave()` to end in `.pdf` instead of `.png` will change the type. Looking at the function documentation, we see there are options for: "eps", "ps", "tex" (pictex), "pdf", "jpeg", "tiff", "png", "bmp", "svg" and "wmf" (windows only).
## Common Problems {-}
Mistakes happen. To everyone.
Even this guy! 
Maybe it's a mismatched `()` or `""`.
Or a `+` in the wrong spot.
But more than likely, you misspelled something.
[](https://allisonhorst.com/everything-else)
[Art by Allison Horst](https://allisonhorst.com/everything-else/)
- Read the error message for hints
- Google error message
- Look up documentation
- Ask R friends
- **Don't give up ... setbacks are temporary**
## Summary {-}
<h3> TADA! </h3>
<h3> Things you now know how to do: </h3>
:::: {style="display: flex;"}
::: {}
- Produce a simple plot with `ggplot2`
- Use different **`aes`thetic mappings** and **`geom_*()` functions** to produce more complex plots
- Visualize **distributions and relationships** in data
- Produce **small multiples** with `facet()`
- [Make the plot to the right](https://r4ds.hadley.nz/data-visualize_files/figure-html/unnamed-chunk-7-1.png)
:::
::: {}

:::
::::
### Resources: {-}
- [R4DS book (1e)](https://r4ds.had.co.nz/data-visualisation.html)
- [R for Data Science: Exercise Solutions (1e)](https://jrnold.github.io/r4ds-exercise-solutions/)
- [A Layered Grammar of Graphics](http://vita.had.co.nz/papers/layered-grammar.pdf)
- [{ggplot2}](https://cran.r-project.org/web/packages/ggplot2/index.html)
- [ggplot2 extensions - gallery](https://exts.ggplot2.tidyverse.org/gallery/)
- [R Graphics Cookbook](https://r-graphics.org/)
- [Gina Reynolds' ggplot flipbook](https://evamaerey.github.io/ggplot_flipbook/ggplot_flipbook_xaringan.html)
## Meeting Videos {-}
### Cohort 5 {-}
`r knitr::include_url("https://www.youtube.com/embed/0NdiQuuM0vw")`
<details>
<summary> Meeting chat log </summary>
```
00:11:14 Jon Harmon (jonthegeek): dslc.io/r4ds
00:12:06 Saeed Shafiei Sabet: Hi everyone!
00:12:29 Sandra Muroy: Hi Saeed!
00:13:05 Becki R. (she/her): Hello!
00:13:30 Sandra Muroy: Hi Becki!
00:22:41 Saeed Shafiei Sabet: Can also by using ggplot2 do some 3D surface plots?
00:24:01 shamsuddeen: https://ggplot2.tidyverse.org/reference/geom_contour.html
00:24:12 shamsuddeen: 2D contours of a 3D surface
00:25:34 Saeed Shafiei Sabet: Thanks @shamsuddeen ;)
00:25:50 Jon Harmon (jonthegeek): ggplot2 is 2D. There are other packages for 3D visualization, I'll try to link some in your question on the Slack once we're done!
00:26:29 Saeed Shafiei Sabet: @Jon Thanks a lot! :)
00:26:34 shamsuddeen: I guess this package provides 3D plotting https://www.rayshader.com/index.html
00:27:24 Jon Harmon (jonthegeek): Yup, that's the one I was going to recommend: https://cran.r-project.org/web/packages/rayshader/index.html
00:28:28 Jon Harmon (jonthegeek): I found it super helpful to figure out how to read some of these things as words:
%>% = "and then"
~ = "by" (usually)
00:28:30 shamsuddeen: Looks at some practical examples of the package here: https://www.tylermw.com/3d-ggplots-with-rayshader/
00:29:54 Saeed Shafiei Sabet: Thank you!
00:36:02 docksbox@pm.me: https://jrnold.github.io/r4ds-exercise-solutions/
00:40:57 Jon Harmon (jonthegeek): ?ggplot2::mpg will show all the details of the dataset
00:41:50 Sandra Muroy: thanks Jon :)
00:42:40 Jon Harmon (jonthegeek): hwy = "highway miles per gallon", cty = "city miles per gallon" in that set, so usually that's what you'd want on y.
00:43:38 Becki R. (she/her): Did I hear correctly that the dependent variable goes on the y-axis?
00:44:04 Jon Harmon (jonthegeek): Generally, yes. But it's whatever you specify as "y" in the "aes" call.
00:44:16 Becki R. (she/her): ok thanks
00:49:24 Jon Harmon (jonthegeek): The "labs" function is for all of the labels for your plot.
00:51:26 Jon Harmon (jonthegeek): https://twitter.com/search?q=%23tidytuesday&src=typed_query
00:51:48 Hector: Is there any specific use for the ggtitle() function in contrast with labs() ?
00:52:09 Njoki Njuki Lucy: what is there a difference between stat="count" and stat="identity"? I understand stat = "count returns count per each level.
00:53:23 Jon Harmon (jonthegeek): @Hector: ggtitle is equivalent to labs() for just the title and subtitle parts. It's just to make it easier to focus on those specific bits.
00:53:56 Jon Harmon (jonthegeek): @Njoki: "count" means "how many entries have this value?", vs "identity" means "what value is in this cell?"
00:54:59 Hector: Thank you!
00:55:04 Njoki Njuki Lucy: thank you.
00:56:26 Jon Harmon (jonthegeek): "color" = outside, "fill" = inside
00:58:53 docksbox@pm.me: labs()
01:04:36 Ryan Metcalf: Could it be stated that “labs” is a more eloquent way of labeling than explicitly calling each field directly? Less lines of code maybe?
01:05:47 Jon Harmon (jonthegeek): I'm not sure I'd say "eloquent," but it's just another option. They provide the separate functions in case you're looking for them, basically.
01:07:23 Jon Harmon (jonthegeek): Sorry about that!
01:08:23 Susie Neilson: This was a great presentation - thank you so much Federica!
01:12:31 docksbox@pm.me: example would be the use of a map data
01:12:51 Saeed Shafiei Sabet: Thanks Federica :)
01:13:43 docksbox@pm.me: great thanks!
01:13:46 Fodil: thank you everyone was very interesting.
01:13:51 Becki R. (she/her): Thanks, Federica!
01:13:56 Njoki Njuki Lucy: Thank you.
01:13:56 Saeed Shafiei Sabet: Thank you
01:13:58 Saeed Shafiei Sabet: bye
```
</details>
### Cohort 6 {-}
`r knitr::include_url("https://www.youtube.com/embed/5IMg8AC-1IA")`
<details>
<summary> Meeting chat log </summary>
```
00:16:33 Daniel Adereti: do I have to run library(tidyverse) everytime I start my R?
00:17:15 Zaynaib Giwa, @zaynaib: You only need to call it once when you open R Studio
00:21:14 Shannon: To double check that you have it installed, you can click on the 'Packages' tab in the lower right pane. If it's installed, you'll see it listed next to a checkbox. Once you load the package with the library() function, you will see a checkmark in the box.
00:28:20 Freya Watkins (she/her): displ 's class is dbl (double) which is a floating point number for decimal precision. int (integer) is for full rounded values (I think)
00:28:49 Zaynaib Giwa, @zaynaib: cute cat
00:28:49 Vrinda Kalia: Yes, I was just typing that up @Freya I think you are right
00:30:26 Aalekhya Reddam: In Line 88 is it a - or = between data and mpg?
00:30:57 Matthew Efoli: I think it is an equal to sign
00:30:58 Aalekhya Reddam: Sorry that was my screen lagging!
00:38:43 Shannon: Another option for finding number of rows and number of columns, that I used, is nrow(mpg) and ncol(mpg). Not as much info as glimpse(), but more concise.
00:41:42 Freya Watkins (she/her): @Shannon, dim(mpg) also gives both dimensions in one command (number of rows, followed by number of columns) :)
00:42:35 Shannon: @Freya, perfect! thanks!
00:49:45 Zaynaib Giwa, @zaynaib: I have to leave a bit early. But it was nice meeting everyone. See you next week.
00:50:02 Shannon: See you next week!
00:50:30 Matthew Efoli: See you next week Zaynaib
00:50:34 Aalekhya Reddam: I have to head out too, see you all next week! :)
00:50:57 Shannon: 👋
00:51:06 Matthew Efoli: Have a nice weekend Aalekhya!
00:51:20 Freya Watkins (she/her): missing the closing bracket I think
00:53:58 Daniel Adereti: See you next week Aalekhya and Zaynaib!
00:54:27 Daniel Adereti: We have 10 more minutes, so we may have to carry over the chapter to next week, I think
00:55:10 Matthew Efoli: @Daniel okay
00:56:31 Shannon: That works for me. I'd have more time to look over the second half of the chapter. :)
00:56:52 Marielena Soilemezidi: Yes, same for me! :)
00:56:52 Vrinda Kalia: Yes, this sounds good to me as well!
00:59:17 Shannon: Nice work presenting, Adeyemi! Thank you!
00:59:20 Marielena Soilemezidi: Daniel, could you share the link of the spreadsheet here?
00:59:40 Daniel Adereti: https://docs.google.com/spreadsheets/d/1zy2nXNkvcdqWuF8rQ5ApWRkVQG_UJt0azu3h_mEnY2E/edit#gid=0
00:59:43 Marielena Soilemezidi: Because I couldn't find it in Slack for some reason
00:59:47 Marielena Soilemezidi: thank you!!
01:00:35 Daniel Adereti: 👍
01:08:40 Vrinda Kalia: Maybe try “stroke = class” ?
01:10:01 Freya Watkins (she/her): try stroke = 5. it should modify the width of the border so I think needs a numeric argument
01:10:08 Daniel Adereti: maybe this? ggplot(data = mpg)+
geom_point(mapping = aes(x = displ, y = hwy),
shape = 17,
stroke = 3)
01:10:32 Vrinda Kalia: I see, it needs a numeric argument
01:11:37 Vrinda Kalia: Thank you so much for leading the discussion, Adeyemi!
01:11:55 Matthew Efoli: Thank you so much Adeyemi!
01:11:56 Freya Watkins (she/her): Thank you, Adeyemi! Great job :)
01:11:57 anacuric: thank you!
01:11:58 Marielena Soilemezidi: Thank you guys! And thanks Adeyemi for presenting!! :)
01:12:00 Shannon: Thank you everyone!
01:12:01 Adeyemi Olusola: thank you
01:12:02 Marielena Soilemezidi: See you next week!
01:12:16 Matthew Efoli: see you next week!
```
</details>
`r knitr::include_url("https://www.youtube.com/embed/sEYMNIEvG5Q")`
<details>
<summary> Meeting chat log </summary>
```
00:06:29 Adeyemi Olusola: Good day Daniel
00:06:58 Daniel Adereti: Hello Olusola!
00:07:06 Daniel Adereti: Looking forward to today!
00:07:21 Adeyemi Olusola: Yeah yeah.
00:12:52 Amélie Gourdon-Kanhukamwe: Hi everyone, I am in my shared office at work, so cannot really speak (have been meaning to join from the start, but shared office = loads of distraction). (I am based in London UK).
00:14:12 Shannon: Hi Amelie, nice to 'meet' you :)
00:20:49 Aalekhya Reddam: I may have missed it but what is the difference between adding a period and not adding one for facet_grid?
00:22:41 Shannon: I think I missed that too, I second that question.
00:26:04 Freya Watkins (she/her): The point should have removed one of the dimensions - either vertical or horizontal
00:29:57 Shannon: So, having a point or no point would have the same result? Ex: ( ~ drv) is the same as( . ~drv)? Or (drv ~) is the same as (drv ~ .) ?
00:32:11 Aalekhya Reddam: That’s what I understood too Shannon! The location of the ~ determines orientation and I guess the “.” is a placeholder
00:32:48 Shannon: I think that's how I'm understanding it, too
00:39:09 Freya Watkins (she/her): Yes I believe that's right @Shannon, @Aalekhya - I think in the previous plot there was an extra dimension (drv ~ cyl) which had rows and columns, then replacing drv with a point (. ~ cyl) just returns a plot faceted with columns for cyl. Then (~ cyl) returns the same plot as (. ~ cyl)
00:39:44 Aalekhya Reddam: Ah okay, thank you Freya!
00:39:52 Shannon: Okay, thanks Freya!
00:58:00 Shannon: That last one drove me crazy, I couldn't figure it out! I tried position = jitter and tied to adjust stroke. Looks like the solution is two geom_point layers. Thanks for solving that one!
01:10:45 Aalekhya Reddam: I have another meeting and have to head out, thank you for a great lesson Adeyemi! See you all next week
01:10:57 Vrinda Kalia: Thank you so much, Adeyemi! I appreciate your thoroughness. I need to leave for a 1pm call. See you all next week!
01:11:06 Shannon: See you next week!
01:11:13 Daniel Adereti: Hello guys, will we like to conclude the chapter ourselves next time so we move to the next chapter with Matthew?
01:11:20 Shannon: Thank you Adeyemi!
01:11:30 Marielena Soilemezidi: See you guys! Thank you Adeyemi!!
01:11:32 Matthew Efoli: Thank you Adeyemi
01:11:35 Freya Watkins (she/her): Thanks Adeyemi! Really helpful, thanks for all the extra time and effort on the exercises
01:11:42 Amélie Gourdon-Kanhukamwe: Thank you!
01:11:58 Freya Watkins (she/her): I'm happy to finish the rest myself and move on to next chapter next time :)
01:12:43 Matthew Efoli: I have to go. I will be taking the next chapter
01:12:47 Freya Watkins (she/her): Bye! Thanks Daniel
01:12:52 Matthew Efoli: bye
```
</details>
### Cohort 7 {-}
`r knitr::include_url("https://www.youtube.com/embed/eqQJ5tmEGco")`
<details>
<summary> Meeting chat log </summary>
```
00:08:24 Oluwafemi Oyedele: Let wait for about 5 minute for others to join
01:02:00 Aditi S: Thank you! I have to leave now but hope to join again next week.
01:05:04 Oluwafemi Oyedele: geom_jitter is very useful if you want to avoid over-plotting
```
</details>
### Cohort 8 {-}
`r knitr::include_url("https://www.youtube.com/embed/RRxtbv5QCA4")`
<details>
<summary> Meeting chat log </summary>
```
00:11:18 shamsuddeen: https://www.youtube.com/playlist?list=PL3x6DOfs2NGhS_PhklqT6PwK1Fh7blgP2
00:12:09 Mary Emeraghi: Hello everyone
00:13:16 Mary Emeraghi: Hello Shamsuddeen. Please kindly share the link to the GitHub note page
00:13:41 shamsuddeen: https://happygitwithr.com/
00:13:48 shamsuddeen: https://www.youtube.com/playlist?list=PL3x6DOfs2NGhS_PhklqT6PwK1Fh7blgP2
00:14:22 shamsuddeen: https://r4ds.github.io/bookclub-r4ds/
01:24:50 Bidemi Agbozuadu: Thank you very much! Nice presentation
01:25:32 shamsuddeen: https://docs.google.com/spreadsheets/d/1reByMPb5Og3OHbRgplzV5Jkz_o6CkEYz_adBGfHTFwg/edit#gid=0
```
</details>